A variance partitioning perspective of assortative mating: Proximate mechanisms and evolutionary implications
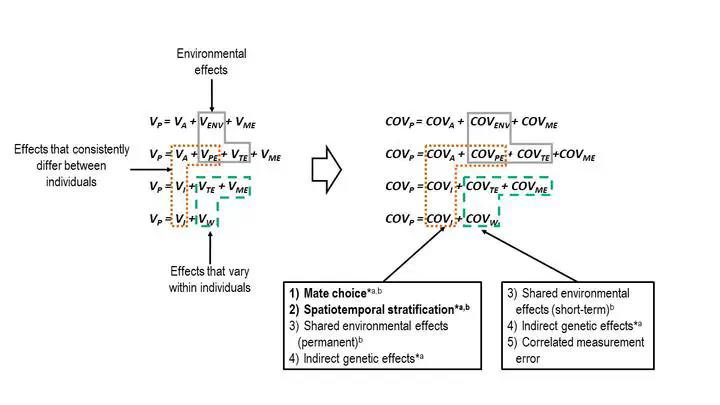 Equations describing the components of phenotypic variance (left) and covariance between pairs (right). Phenotypic variance can be partitioned into different components to which various mechanisms contribute.
Equations describing the components of phenotypic variance (left) and covariance between pairs (right). Phenotypic variance can be partitioned into different components to which various mechanisms contribute.Abstract
Assortative mating occurs when paired individuals of the same population are more similar than expected by chance. This form of non-random assortment has long been predicted to play a role in many evolutionary processes because assortatively mated individuals are assumed to be genetically similar. However, this assumption may always hold for labile traits, or traits that are measured with error. For such traits, there is a variety of proximate mechanisms that can drive phenotypic resemblance between mated partners that, notably, have very different evolutionary repercussions. Bettering our understanding of the role of assortative mating in evolution will thus require insight into its proximate causes. To date, empirical research remains sparse, especially when for labile traits. This special issue aims to stimulate such research while promoting the usage and development of statistical approaches allowing the quantification of the relative roles of alternative proximate mechanisms causing assortative mating. To this end, we first describe how the phenotypic covariance between mated partners can be usefully partitioned into components that capture one or several of five distinct mechanisms. We then demonstrate why the importance of mechanisms causing genetic covariance between the traits of partners may often be overestimated. Finally, we detail how the evolutionary causes and consequences of the diverse mechanisms may be identified.